I've been doing some math to help me think about breeding strategies with tomatillos. Last week I showed some code for calculating how populations of different sizes converge under selection for a single recessive trait. Here I'll show similar code for a single dominant trait.
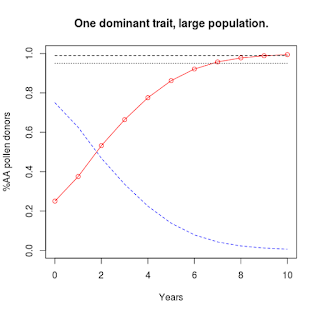 |
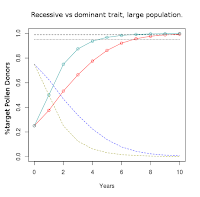
Solid red curve with circles: %AA pollen donors.
Dashed blue curve: %Aa & %aa pollen donors. |
Like before, we'll start with an infinite population.
Since we can't tell the difference between plants with one or two copies of the dominant trait ("AA" or "Aa"), we can't tell what the genetic status is of any one plant that we save seeds from. Our goal is a population entirely consisting of "AA" plants, so that is what the code will plot.
The zero year is our F2 population. It takes seven years for the "AA" individuals to represent 95% (dotted horizontal line) of the population. Three years later the level crosses above 99% (dashed horizontal line) of the population.
Because this is the infinite population scenario, there will always be a small percentage of the population carrying the recessive allele.
R Script 3: One dominant trait, infinite population.
# One dominant trait, infinite population.
# Stabilize progeny for dominant trait via selection.
# Save seeds from dominant plants each generation.
years <- 10;
# Define F2 population.
P_AA <- vector();
P_Aa <- vector();
P_aa <- vector();
P_AA <- 0.25;
P_Aa <- 0.50;
P_aa <- 0.25;
# Save seeds only from (AA and Aa) plants, unknown pollen donor. Iterate over years.
for(i in 1:years) {
P_AA <- append(P_AA, P_AA[i]*P_AA[i]*1.00 + P_AA[i]*P_Aa[i]*0.50 + P_Aa[i]*P_Aa[i]*0.25);
P_Aa <- append(P_Aa, P_AA[i]*P_aa[i]*1.00 + P_AA[i]*P_Aa[i]*0.50 + P_Aa[i]*P_aa[i]*0.50 + P_Aa[i]*P_Aa[i]*0.50);
P_aa <- append(P_aa, 0);
P_sum <- P_AA[i+1] + P_Aa[i+1];
P_AA[i+1] <- P_AA[i+1]/P_sum;
P_Aa[i+1] <- P_Aa[i+1]/P_sum;
}
# Make figure.
plot( 0:years, P_AA, col="red", main="One dominant trait, large population.", xlab="Years", ylab="%AA pollen donors", xlim=c(0,years), ylim=c(0,1), axes=TRUE, frame.plot=TRUE);
lines(0:years, P_AA, col="red");
lines(0:years, P_Aa+P_aa, col="blue", lty="dashed");
lines(c(0,years),c(0.95,0.95), col="black", lty="dotted");
lines(c(0,years),c(0.99,0.99), col="black", lty="dashed")
 |
Cyan line w/circles: recessive selection.
Red line w/circles: dominant selection. |
To compare the trajectory for selection on the recessive allele vs on the dominant allele, I overlaid the two curves in an image editor. I inverted the colors for the recessive curves to better distinguish them from the added dominant curves.
Selection on a dominant trait progresses at a slower rate initially than selection on a recessive trait, but by about ten years the two approaches would be expected to reach a similar degree of completeness.
With smaller population sizes, we'd expect the selected allele (dominant or recessive) to reach complete saturation by about that time point.
With recessive traits, I only had to consider "aa" plants as seed producers. With dominant traits, I have to consider "AA" and "Aa" plants. This seems like a small difference, but for simulating small numbers this adds significant complexity.
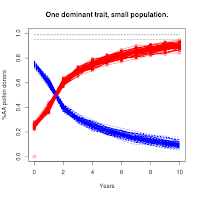 |
| Population = 1000 |
|
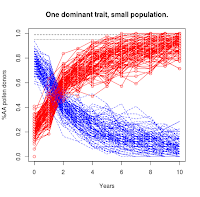 |
| Population = 50 |
|
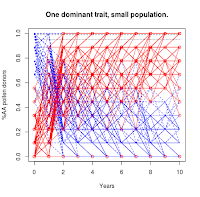 |
| Population = 10 |
|
If you compare these plots to those for the recessive selection scenario (
https://the-biologist-is-in.blogspot.com/2020/01/tomatillo-breeding-2n.html), you'll see that this scenario has a much higher level of noise in the trajectories. For the smallest population level, it takes 30 years (not shown in figures) for the majority of the experimental replicates to converge on the targeted "AA" condition.
R Script 4: One dominant trait, small population.
# One dominant trait, small population.
# Stabilize progeny for dominant trait via selection.
# Save seeds from dominant plants each generation.
years <- 10;
population <- 1000; # 1000, 50, 10
trials <- 100;
# Intialize figure.
plot( c(0,years),c(0,years), col="red", main="One dominant trait, small population.", xlab="Years", ylab="%AA pollen donors", xlim=c(0,years), ylim=c(0,1), axes=TRUE, frame.plot=TRUE);
lines(c(0,years),c(0.95,0.95), col="black", lty="dotted");
lines(c(0,years),c(0.99,0.99), col="black", lty="dashed");
for (ii in 1:trials) {
# Define F2 population probabilities for selection on AA plants.
P_AA_1 <- vector();
P_Aa_1 <- vector();
P_aa_1 <- vector();
P_AA_1 <- 0.25;
P_Aa_1 <- 0.50;
P_aa_1 <- 0.25;
# Define F2 population probabilities for selection on Aa plants.
P_AA_2 <- vector();
P_Aa_2 <- vector();
P_aa_2 <- vector();
P_AA_2 <- 0.25;
P_Aa_2 <- 0.50;
P_aa_2 <- 0.25;
# Save seeds only from (AA and Aa) plants, which can't self-polinate.
for (i in 1:(years+2)) {
# Generate actual population.
rands <- runif(population, 0, 1);
Genotypes <- vector();
for (j in 1:population) {
if (rands[j] < P_AA_1[i]) {
Genotypes <- append(Genotypes, "AA");
} else if (rands[j] < P_AA_1[i]+P_Aa_1[i]) {
Genotypes <- append(Genotypes, "Aa");
} else {
Genotypes <- append(Genotypes, "aa");
}
}
Genotype_counts <- table(Genotypes);
# Determine actual genotype probabilities for pollen donors. (Assuming "AA" plant in case 1, "Aa" plant in case 2.)
if (is.na(Genotype_counts["AA"])) {
P_AA_1[i] <- 0;
P_AA_2[i] <- 0;
} else {
P_AA_1[i] <- (Genotype_counts["AA"]-1)/(population-1); # The plant we're saving seeds from can't be polinated by itself.
P_AA_2[i] <- Genotype_counts["AA"]/(population-1);
}
if (is.na(Genotype_counts["Aa"])) {
P_Aa_1[i] <- 0;
P_Aa_2[i] <- 0;
} else {
P_Aa_1[i] <- Genotype_counts["Aa"]/(population-1);
P_Aa_2[i] <- (Genotype_counts["AA"]-1)/(population-1); # The plant we're saving seeds from can't be polinated by itself.
}
if (is.na(Genotype_counts["aa"])) {
P_aa_1[i] <- 0;
P_aa_2[i] <- 0;
} else {
P_aa_1[i] <- Genotype_counts["aa"]/(population-1);
P_aa_2[i] <- Genotype_counts["aa"]/(population-1);
}
# Generate new theoretical genotype probabilities.
P_AA_1 <- append(P_AA_1, P_AA_1[i]*P_AA_1[i]*1.00 + P_AA_1[i]*P_Aa_1[i]*0.50 + P_Aa_1[i]*P_Aa_1[i]*0.25);
P_Aa_1 <- append(P_Aa_1, P_AA_1[i]*P_aa_1[i]*1.00 + P_AA_1[i]*P_Aa_1[i]*0.50 + P_Aa_1[i]*P_aa_1[i]*0.50 + P_Aa_1[i]*P_Aa_1[i]*0.50);
P_aa_1 <- append(P_aa_1, 0);
P_AA_2 <- append(P_AA_2, P_AA_2[i]*P_AA_2[i]*1.00 + P_AA_2[i]*P_Aa_2[i]*0.50 + P_Aa_2[i]*P_Aa_2[i]*0.25);
P_Aa_2 <- append(P_Aa_2, P_AA_2[i]*P_aa_2[i]*1.00 + P_AA_2[i]*P_Aa_2[i]*0.50 + P_Aa_2[i]*P_aa_2[i]*0.50 + P_Aa_2[i]*P_Aa_2[i]*0.50);
P_aa_2 <- append(P_aa_2, 0);
P_sum_1 <- P_AA_1[i+1] + P_Aa_1[i+1];
P_AA_1[i+1] <- P_AA_1[i+1]/P_sum_1;
P_Aa_1[i+1] <- P_Aa_1[i+1]/P_sum_1;
P_sum_2 <- P_AA_2[i+1] + P_Aa_2[i+1];
P_AA_2[i+1] <- P_AA_2[i+1]/P_sum_2;
P_Aa_2[i+1] <- P_Aa_2[i+1]/P_sum_2;
# Weighted average of the two probability sets by proportion of "AA" vs "Aa" plants.
# Only _1 values carry over to next iteration.
if (is.na(Genotype_counts["AA"])) {
count_AA <- 0; } else {
count_AA <- Genotype_counts["AA"];
}
if (is.na(Genotype_counts["Aa"])) {
count_Aa <- 0; } else {
count_Aa <- Genotype_counts["Aa"];
}
weight1 <- count_AA/(count_AA+count_Aa);
weight2 <- 1-weight1;
val_AA_1 <- P_AA_1[i+1];
val_AA_2 <- P_AA_2[i+1];
val_Aa_1 <- P_Aa_1[i+1];
val_Aa_2 <- P_Aa_2[i+1];
P_AA_1[i+1] <- val_AA_1*weight1 + val_AA_2*weight2;
P_Aa_1[i+1] <- val_Aa_1*weight1 + val_Aa_2*weight2;
if (is.na(P_AA_1[i+1]) == TRUE) { P_AA_1[i+1] <- 0; }
if (is.na(P_Aa_1[i+1]) == TRUE) { P_Aa_1[i+1] <- 0; }
if ((P_AA_1[i+1]+P_Aa_1[i+1]) == 0) {
# End simulation cycle if no "AA" or "Aa" plants.
for (j in (length(P_aa_1)):years) {
P_AA_1 <- append(P_AA_1, 0);
P_Aa_1 <- append(P_Aa_1, 0);
P_aa_1 <- append(P_aa_1, 0);
}
break;
}
## Debugging output.
#message("Iteration ", i);
#print(Genotypes);
#message(" ");
}
# Add current simulation cycle to figure.
points(0:years, P_AA_1[1:(years+1)], col="red");
lines( 0:years, P_AA_1[1:(years+1)], col="red");
lines( 0:years, 1-P_AA_1[1:(years+1)], col="blue", lty="dashed");
}
This essentially means it isn't possible to selectively breed a dominant trait to complete saturation in a small population just using simple selection.
Unlike in the recessive case, we can't just save a few plants over winter to reset the population with only the exact genetics we want. A similar strategy should allow for more rapid progress towards the goal, however.
I'll explore this topic further next time.